Taiji
The Taiji software is an integrative multi-omics data analysis framework. It can be used as a standalone pipeline to analyze ATAC-seq, RNA-seq, single cell ATAC-seq or Drop-seq data. However, the uniqueness and the power of Taiji really lie in its ability to integrate diverse datasets and use these information in a clever way to construct regulatory network and identify candidate driver genes.
The design philosophy of the Taiji pipeline is focused on:
- Correctness: We only include reliable algorithms and make every effort to ensure the implementations are bug-free.
- Performance: We code algorithms from scratch when necessary to ensure the pipeline can scale to large datasets (thousands of samples at least).
- Convinence: Most analyses have multipe entry points, e.g., Fastq, Bam or Bed. The execution of the pipeline requires only a single command.
We achieve these at the expense of customization. This will be improved in the future.
Quick Start
File 1: config.yml
input:input.tsv
output_dir:
output/
assembly:
GRCh38
File 2: input.tsv
| type | id | group | rep | path | tags |
| ATAC-seq | control | control | 1 | ENCFF893KQZ,ENCFF443HVZ ENCODE | |
| ATAC-seq | 2h | 4h | 1 | ENCFF173INV,ENCFF322IZC ENCODE | |
| ATAC-seq | 4h | 4h | 1 | ENCFF562JHD,ENCFF943SYH ENCODE |
taiji run -- config config.yml -n 3 +RTS -N3
Tutorials
If you have used Taiji in your research, please condier citing the following paper:
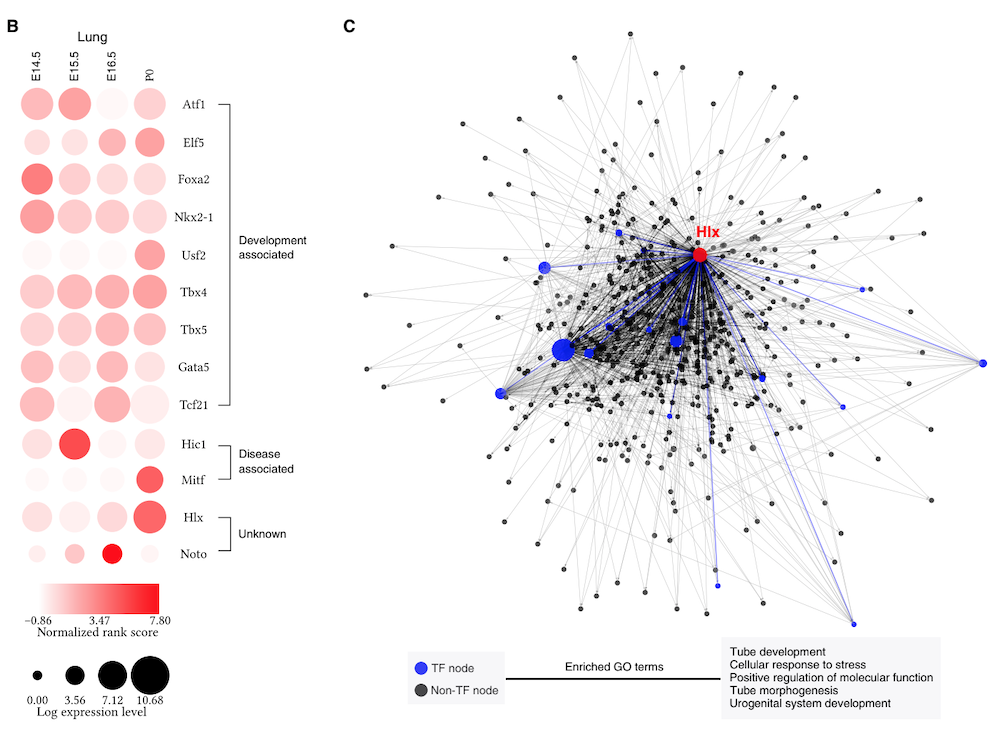
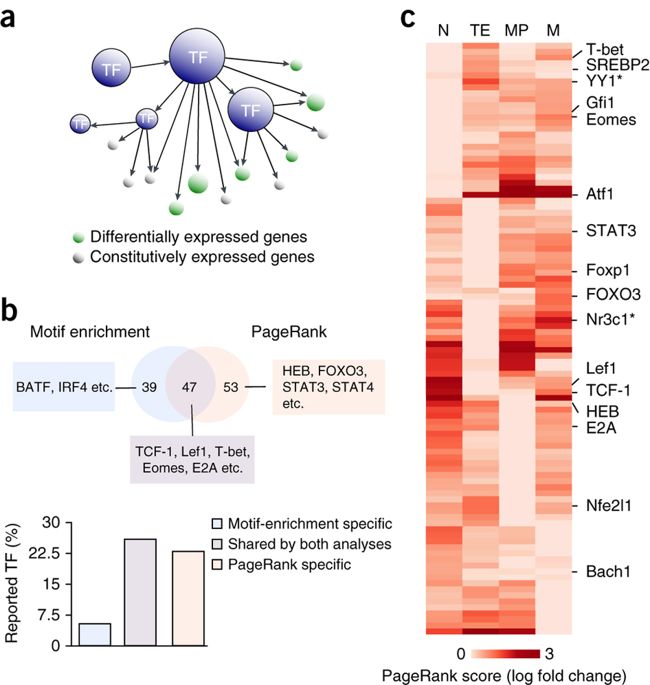
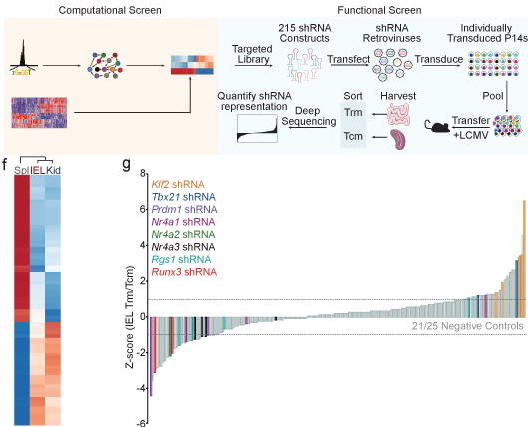
K. Zhang, M. Wang, Y. Zhao, W. Wang, Taiji: System-level identification of key transcription factors reveals transcriptional waves in mouse embryonic development.
Sci. Adv. 5, eaav3262 (2019).